Application-driven MRI
Summary
The traditional imaging pipeline for clinical information extraction (acquisition, reconstruction, analysis) is prone to error propagation and inefficiencies. In cases where the information looked for in an MR image is known beforehand, data processing could be adaptive such that relevant clinical information is obtained directly and reliably. This opens up new possibilities and adds flexibility to the way raw MR data is handled, leading to more efficient paradigms for information extraction from medical imaging data.
Problem and objectives
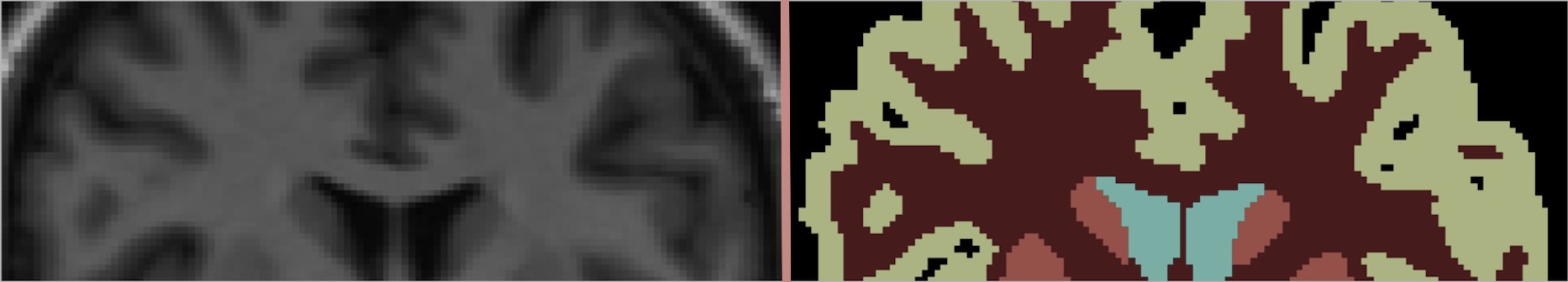
As a particular example of the wider concept of Application-driven MRI, we focus on the problem of joint reconstruction-segmentation of undersampled MR data. We apply this method for left ventricle segmentation in cardiac cine data and for brain tissue segmentation in brain MR. MR acquisition is notoriously time-consuming, and pixel intensity images are rarely the end goal of an MR image. It is usually through further analysis of acquired images that clinically relevant information is extracted. The segmentation of the left ventricle in cardiac cine scans provides invaluable information about cardiac function and mass, revealing clinical indices such as ejection fraction. Likewise, brain tissue segmentation informs brain disseases diagnosis and analysis.
Joint reconstruction and segmentation
We propose the modification of a compressed sensing reconstruction method with the introduction of a discriminative model for the image. This forces intensity pixels to be estimated consitently with data acquired and simultaneously belong to one of a few segmentation classes. The discriminative model used is a mixture of Gaussians, therefore penalising intensity values that deviate too much from the mean of the Gaussian they have been attributed. Clusters of pixel intensities are formed and pulled apart, enhancing edges between different regions of the intensity image.
Results
The joint reconstruction-segmentation is compared against a Gaussian mixture model segmentation of the fully sampled data. As an alternative workflow to obtain segmented MR images from undersampled data, we compare the performance with a separate method that first reconstructs an intensity image using compressed sensing and then segments it in a separate stage. Although changes in the intensity image reconstruction are subtle, we are able to obtain up to 30% less left ventricle pixel misclassification using the joint approach. Brain tissue segmentation is also shown to be more accurate than the separate method relative to manual labelling. The use of the discriminative modelling then has two consequences: first, it allows to obtain a segmentation of the image as a by-product of the reconstruction, and also, the joint approach better conditions the data for its segmentation.